
Recent organ-on-a-chip advances toward drug toxicity testing
Introduction
Since 2011, when US President Obama announced the establishment of the “Microphysiological System” research project, “organ-on-a-chip” systems have experienced a research boom around the world. These systems integrate microengineering, microfluidics, and biomimetic principles to create bionic devices that mimic the primary functions of human organs within a tiny volume (1). The dimensions of the on-chip components make it possible to accommodate molecules, cells, tissues, and even organs at the same time, and the chip’s special fluid-handling control system enables it to simultaneously measure physical quantities, chemical quantities, and biomass (2). In addition to the advantages of miniaturization, integration, and low consumption of fluids, organ chip technologies can accurately control multiple system parameters, such as chemical concentration gradients (3), fluid shear forces (4), the construction of cell-based graphic cultures (5), and tissue organization (6). Interfaces and organ-organ interactions (7) can be used to simulate the complex structures, microenvironments, and physiology of human organs. To date, many organ-on-a-chip models have been established for the study of organ-level biology and functions, disease mechanisms, and screening for drugs.
Drug toxicity is the most frequent cause of early termination of clinical trials, and can even lead to the post-market withdrawal of drugs, resulting in the loss of human and financial resources. Lack of efficient drug toxic predicting models lead to insufficient insight into the toxic mechanism for preclinical outcomes (8). Many results have indicated that the organ-on-a-chip technology is effective for bridging the gap between preclinical and clinical outcomes. Further, multi-organ-on-chip integrated several artificial organs via recirculation fluidic flow, which create highly functional and robust systems and enable a more accurate model to drug testing (9-11). In this review, we introduce some recent advances in the novel organ-on-a-chip studies that have been applied to assessing drug toxicity. Especially, we put the emphasis on the heart-, liver-, kidney-, and brain-on-a-chip. These four organs are the main targets of the drug toxicity. Furthermore, we discuss a number of representative multi-organ-on-a-chip researches in the recent for drug toxicity testing.
Heart-on-a-chip
Cardiomyocytes, a type of cross-striated muscle cells, are contractile cells that allow the heart to pump. Physiologically, the majority of the cardiomyocytes are aligned with their long axes parallel to the epicardial ventricular surface of the heart. Cardiomyocytes attach to each other using gap junctions and fascia adherens unions to form the myocardial fibers, which are responsible for contraction. Recently, microfluidic heart-on-a-chip systems were developed to mimic the structure and function of the myocardium. For example, muscular contraction and electrical activity are the most important characteristics of cardiomyocytes. To measure muscular contraction, elastic thin-films are commonly employed. Based on muscular thin-film technology, Grosberg et al. designed a heart-on-a-chip that can measure contractility (Figure 1A) (12). A high-throughput heart-on-a-chip was developed by Agarwal et al. Thin-film polydimethylsiloxane (PDMS) cantilevers was used to mimic the laminar architecture of the heart ventricle. The diastolic and systolic stresses were reproduced by the biomimetic organ (13). Muscular thin-film cantilevers were developed by McCain et al. using gelatin hydrogel. Myocytes on gelatin had higher spare respiratory capacity than those on PDMS coated with fibronectin (14). Nawroth et al. reported that UV-patterning shortened the time for gelatin substrate fabrication, compared with common used lithography. This method facilitates continuous automated construction of heart-on-a-chip (15). To measure the electrical activity and force of muscle contraction, Stancescu et al. integrated separate two-dimensional (2D) cellular systems with Biomicroelectromechanical Systems (BioMEMS) constructs. Microelectrodes embedded in extracellular matrix could be used to measure the electrical properties of 2D cultured cardiac cell (16). Lind et al. developed some cantilevers that contained flexible thin-film strain gauges. Cardiac muscle tissue was cultured on these cantilevers (Figure 1B) (17). Beat rates and contractile stresses of the cardiac cells could be detected by the flexible strain gauge. Tanaka et al. aimed to develop a microfluidic device without the need for external energy sources. They created a PDMS hollow sphere powered by spontaneously contracting cardiomyocyte sheets. The devices worked continuously for at least 5 days (18). In contrast with elastic thin-film devices, Sidorov et al. designed an ‘‘I-Wire” platform that interrogated passive and active mechanical and electrical characteristics. A calibrated flexible probe provided the strain load of the engineered three-dimensional (3D) cardiac tissue constructs (ECTC) via lateral displacement. The ECTCs exhibited longitudinally aligned cardiomyocytes with well-developed sarcomeric structure. The “I-Wire” platform enabled the creation, as well as the mechanical and electrical characterization, of ECTCs. Therefore, they can be valuable for the study of cardiac diseases and drug development (19,20).
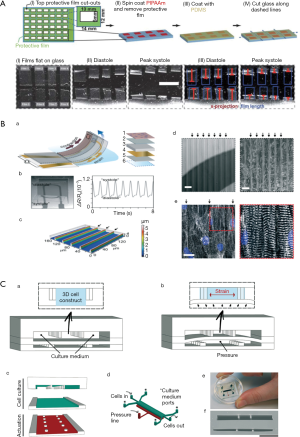
Another research focus is cell differentiation and architecture. For example, some microfluidic models were developed to promote cardiomyocyte differentiation, proliferation, and the formation of myocardial-like tissues. Wang et al. reported that induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) seeded onto thin elastomers with patterned lines of fibronectin self-organized into anisotropic myocardial tissues, called muscular thin-films. Based on this “heart-on-a-chip,” they found that Barth syndrome (BTHS) iPSC cardiomyocytes assembled into irregular and scattered sarcomeres. Weak contraction was detected using the BTHS “heart-on-a-chip” (21).
Marsano et al. reported a “heart-on-a-chip”. Myocardium was used to mimic the physiological and mechanical microenvironment of heart (Figure 1C) (22). Hanging post array was used to confine the gels with embedded cells. Homogeneous uniaxial cyclic strains were generated by a pneumatic actuation system to stimulate the cell-laden gels. Using both rat and human stem cells, functional micro-engineered cardiac tissues (µECTs) were generated. Parsa et al. reported a pneumatic microfluidic platform. Cardiac micro-tissues could be manipulated repetitively over several weeks. This device was used to study cardiac hypertrophy in high throughput (23). Kobuszewska revealed that the geometry of microsystems and microenvironmental influence the arrangement, morphological features, and proliferation of cardiac cells. The proliferation of cardiac cells did not enhance in static conditions. In contrast, perfusion conditions influenced the proliferation and alignment of cardiac cells (24).
Recently, 3D-printing has become an emerging method for in vitro model construction, with the advantage of accurate positioning to help replicate subtle complex structures. For instance, an endothelialized myocardium fabricated by 3D-bioprinting was proposed by Zhang et al. Endothelial cells were bioprinted in microfibrous hydrogel using composite bio-ink. It was found that these cells migrated gradually towards the peripheries of the microfibers and form a confluent layer. Then, cardiomyocytes were seeded with the 3D endothelial bed. An aligned myocardium capable of spontaneous and synchronous contraction was generated. The organoids were further embedded into a microfluidic perfusion system to develop an endothelialized-myocardium-on-a-chip platform (25). Wang et al. tried to construct a contractile cardiac tissue using a 3D-bioprinting strategy. Primary cardiomyocytes from infant rat hearts were used. The bioink is fibrin. Spontaneous synchronous contraction was detected in the bioprinted cardiac tissue. It suggests that cardiac tissue was developed and matured in vitro. Immunostaining for a-actinin and connexin further confirmed the development of cardiac tissue (26).
Liver-on-a-chip
The liver is the largest organ in the abdominal cavity of rodents, has a variety of important functions, and plays a vital role in maintaining homeostasis of the internal environment. These include regulating blood glucose levels, synthesizing various plasma proteins, detoxification, and immune regulation. The biotransformation of exogenous substances, such as drugs, occurs mainly within the liver. As a result, the liver is the most vulnerable organ to foreign substances, and especially their metabolites. Drug induced liver injury has become one of the most important factors that affects the safety of a drug for clinical use.
To date, many animal or in vitro models have been employed to test drug metabolism and inspect hepatotoxicity. However, these models are far from satisfactory due to extensive species differences or rapid dedifferentiation of hepatocytes in 2D-cultures, resulting in the loss of liver phenotype and function. 3D-culture techniques were used to increase cell-cell interactions to create a microenvironment closer to that of the body, and to enhance the specific functions of liver cells in vitro. Zuchowska et al. constructed a high-throughput spheroid culture model to detect the hepatotoxicity of 5-FU, an anticancer drug, using HepG-2 cells (27) (Figure 2A). It was found that the initial size of the spheroids affected the toxicity of 5-FU, and as the diameter of spheroids increased, the drug resistance decreased. In addition, the lack of expression of a variety of key metabolic enzymes and phase II conjugation reactions involved in drug clearance leads to inconsistent results compared with tests conducted in vivo (28). This single-culture method lacks the biochemical cues and intercellular communication between the parenchymal and non-parenchymal cells, which include the liver sinusoidal endothelial cells, hepatic stellate cells, and Kupffer cells. All of these interactions are necessary for maintaining the physiological phenotype of parenchymal cells and investigating the mechanisms of hepatotoxicity (29). Du et al. encapsulated hepatocytes with endothelial cells, differentiating them from hiPSCs produced with recombinant proteins, placed them within specific niches in multicomponent hydrogel fibers, and assembled them into 3D-patterned endothelialized liver tissue constructs (30). In addition, recent research has reported that no single-cell culture model can accurately predict liver toxicity when the pharmacokinetic data are missing, especially using an end-point method, which is suitable for primary cells (31).
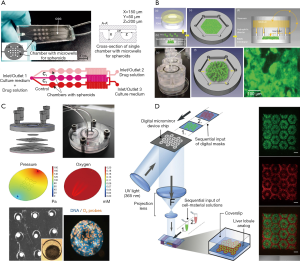
Co-culture hepatocytes with non-parenchymal cells was used to detect hepatotoxicity. Hendriks et al. reported on a 3D-spheroidal model that showed significant polarization characteristics, such as the expression of MRP2 and bile output pump protein (32). This model could be used to detect liver toxicity caused by chronic or multiple exposures. Leite et al. designed scaffold-free organoid spherical culture models in pairs to co-culture hepatocytes with stellate cells (33). The results showed that 3D co-culture models exhibited higher liver function compared with 2D models or 3D models. Their metrics included albumin synthesis, urea secretion, and gene expression characteristic of hepatocytes and stellate cells. This model achieved activation of hepatocyte-associated or drug-related stellate cells for up to 21 days. When exposed to allyl alcohol or methotrexate, stellate cells became activated and extracellular matrix secretion was increased. Thus, the integration of co-cultivation and 3D-cultivation is an important developmental trend for the detection of hepatotoxicity in future in vitro models.
Another feature of the in vivo microenvironment are the shear forces from blood or other fluids. For this purpose, the perfusion culture technique is also applied to the in vitro culturing of some liver-on-a-chip systems to more accurately reflect the physiology and microenvironment in vivo. Rennert et al. inoculated four types of hepatocytes into a porous membrane, and applied perfusion culture conditions to create a biomimetic liver model that was more physiologically accurate (34). The results showed that the presence of shear forces and the ordered spatial arrangement of the cells gave the model more realistic morphological and functional characteristics. This model provided an excellent precedent for follow-up liver toxicity studies based on membrane perfusion systems.
Many drugs do not exhibit hepatotoxicity in the short term, but only after long-term administration or multiple doses, which requires the in vitro model to remain phenotypically stable, as well as keeping its morphology, viability, and hepatocyte-specific functions while maintaining its metabolic capacity during long culture times. An easily scalable 3D human primary hepatocyte spheroid co-culture model was proposed for the study of drug-induced chronic hepatotoxicity, along with liver function and some pathological mechanisms (35). This spherical culture model can be maintained in vitro for 5 weeks while maintaining high CYP450 enzyme activities and albumin secretion, this model successfully detected the hepatotoxicity of various long-term drugs at clinical concentrations. In addition, owing to differences in oxygen partial pressure and nutrient concentrations in the various regions of the liver, the hepatocyte plate function between the central vein and the portal vein segment is not the same, so the reactions to liver toxicity are different. Weng et al. constructed a device so that the local distribution of the liver was reconstructed by the self-assembly of the cells, and the results showed that areas farther from the central vein experienced higher hepatotoxicity (Figure 2B) (36). When a fluorescent probe is added to the chip, the process of liver injury can be simulated in real time. For example, after Bavli et al. embedded fluorescent probes into a liver-on-a-chip system, it responded to drugs in real time, and the mitochondrial function switched from oxidative phosphorylation to glycolysis. This change of energetic mode indicates that the mitochondria became damaged (Figure 2C) (37).
In addition, the relatively high production cost and complexity of existing liver organs limit their large-scale development for toxicity testing. Using 3D-printing technology to automate the high-throughput production of liver organ chips or liver tissue will greatly reduce production costs, and the operation is relatively simple. Bhise et al. used 3D-printing technology to produce long-lasting 3D human HepG2/C3A spheroids for drug toxicity assessment (38). Their report pioneered the use of 3D liver organ chips for toxicity testing. In another example, Ma et al. used a rapid digital 3D-bioprinting technology to construct bionic liver lobule models with higher liver-specific gene expression levels, increased metabolite secretion, and enhanced cytochrome P450 induction (Figure 2D) (5).
Consequently, liver-on-chip systems are of great significance for developing drug hepatotoxicity tests, and for solving the shortcomings of in vitro model models, such as low sensitivity and poor accuracy, as well as the need for animal experiments, complicated experimental procedures, lengthy test cycles, species differences, and ethical issues. Still, many factors remain to be addressed. For example, a liver-on-a-chip system depends on all kinds of hepatocytes and liver micro-tissues. The origin of the cells largely determines the function of the in vitro model, along with the reliability of the results. Human primary hepatocytes are considered to be the gold standard for detecting hepatotoxicity in an in vitro model, but the sources are very limited, and maintaining hepatocyte function in vitro is still a major problem for the scientific community. Stem cells, especially iPS cells, have a theoretically unlimited supply, and can be differentiated into any cell type with special functions, so they have the potential to be developed into individualized drugs and liver toxicity tests. Although the current study of functional cells after iPS differentiation is not complete, this trend shows no signs of stopping. However, no model to date has been able to collect bile, and excessive bile accumulation leads to cholestatic liver toxicity. Models with bile collection systems will more accurately reflect liver toxicity. So far, only in vivo models can handle all aspects of drug distribution in the body, and a single liver organ cannot account for the pharmacokinetic changes of drugs entering the body, such as the absorption of the small intestine, the distribution of the drug in the body, and excretion by the kidneys. It is known that the gut and gut microbiome, along with the kidneys, affect the systemic utilization of drugs and their toxicity. Multi-organ chips suitable for long-term culture will be critical for future development.
Kidney-on-a-chip
The kidney is responsible for generating urine to remove metabolites and certain wastes and toxins from the body. At the same time, the kidney recycles water and other useful substances in its reabsorption function. However, owing to poor prediction of adverse drug effects and insufficient insight into toxic mechanism with preclinical outcomes, 19% of drug attrition occurred in Phase 3 of pharmaceutical development (39). Even in the post approval stage, 20% of acute renal failure in community and hospital settings was caused by drug-induced kidney injury. Differences in drug transporter and metabolizing enzyme expression between species (40), and the lack of similarity between preclinical cell studies in vitro and clinical use is to blame. To bridge the gap between the preclinical and clinical outcomes and improve the predictability of nephrotoxicity, developing new in vitro models for drug-induced kidney injury is a future development trend. The “five similar” qualities are needed for this purpose: similar key functional cell types, functional structures, substance delivery methods, system liquid components, and fluid-dynamics environments. Kidney-on-a-chip systems are considered to have great potential as predictive models.
Current kidney-on-a-chip systems range from simple to complex. With regard to structure, some chips were originally designed as culture systems with cells seeded in the chamber and the culture medium continuously flowing toward the apical sides of tubular epithelial cells (41). Then, a porous membrane is introduced, which causes the basal sides of the cells to be exposed to the culture medium, leading to cell polarity formation. The new chips promote drug transporters expressed at the correct positions of the cells, which was not possible in traditional 2D platforms. The environments of cell culture are more varied, from 2D (42) to 3D (Figure 3A). A 2016 study described a 3D culture system in which extracellular matrix facilitates the formation of tubule structures, and the cells in the systems expressed higher levels of functional protein (43). The types of cells used on the chip also increased from one type to three types (Figure 3B) (44,45). The types of simulated nephron segments were also diversified, including renal tubules (46), collecting tubules (47), and glomeruli. The number of simulated nephron segment types in the same system was also increased from one to two (48). The potential cell sources are widespread, including cell lines (49-51), primary cells (52), and pluripotent stem cell-derived cells. The construction technology has been improved by the application of 3D-bioprinting, and the construction of complex 3D culture systems has become easier. Lewis et al. used human proximal renal tubular epithelial cells to build a 3D chip (Figure 3C) containing a convoluted tubule using a 3D-bioprinting approach. In contrast with other reported kidney-on-a-chip systems based on common PDMS manufacturing methods, the renal tubule on this chip can wreathe flexibly (53).
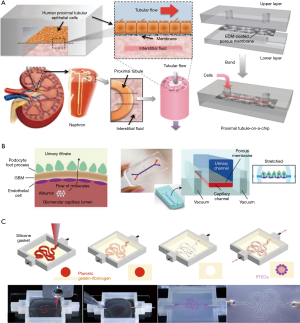
While these works improve upon technology used to restore the function of kidney systems on microfluidic chips, the development of a true kidney-on-a-chip is still in its early stages. The microenvironments mimicked in these chips were comparatively simple. Most of them used only one or two types of renal cells, and could not simulate a completed nephron. The composition of the liquids in the chip systems were simple, and not the same as in vivo. A 2018 study considered these issues and described a nephron system that used renal endothelial cells, podocytes, and renal tubular epithelial cells to mimic the functions of glomeruli and renal tubules, and introduced bovine serum albumin (BSA) into the liquid to distinguish “renal blood flow” from “glomerular filtrate drainage”. This system provided a microenvironment for three types of the cells to survive while maintaining their functions, and crudely mimicked the process of drug elimination by the kidney. However, there remain some limitations to this approach. Notably, the substance exchange areas in the glomeruli and tubules cannot match the corresponding areas in actual nephrons, and the simulation of renal hemodynamics was very simple.
There are three primary factors limiting the development of kidney-on-a-chip systems, including issues with cell sources, materials, and available technologies. With regard to cell sources, cell lines and primary cells are commonly used, but only a few works used pluripotent stem cell-derived cells. Cell lines are convenient to use, but they usually lack functionality, and the types are limited. Freshly isolated primary cells should theoretically be able to perform all functions in vivo, but they easily lose their phenotypes with the passage of time, and cannot be widely used because of a scarcity of donors. Pluripotent stem cell-derived cells have the potential to overcome the limitations of primary cells, but the protocols for the directed differentiation of pluripotent stem cells to renal cells are imperfect, and the functional completeness of these cells has yet to be demonstrated. In addition to addressing cell source limitations, the required materials also need be to be improved. Softer, flexible, and more biocompatible substrates are needed to match the requirements for mimicking renal hemodynamic characteristics. In addition, construction and detection technologies require more development. Because of the tiny sizes of the chips, the sample volumes are small, so many traditional detection methods cannot be used, which limit data diversification and comparisons with in vivo situations.
In the early stages of kidney-on-a-chip development, primary cells from rodents are required for two reasons. First, the cells have robust functions and are sensitive to the microenvironment, so that the cells can serve as sensors to detect the biocompatibility of the system. Second, unlike most data from humoral detection in humans, the data obtained from rodent cells are more diversified, so that more comprehensive comparisons can be made between data from the chip and in vivo tests. In addition, to develop better kidney-on-a-chip systems, the evaluation methods need to be standardized to direct the designs of the systems. In addition, more biomarkers appropriate for both in vitro and in vivo experiments are required to translate the data between situations. Finally, chip models should be made simple or complex based on the specific requirements of the application. The flexible combination of highly integrated kidney-on-a-chip components can be connected as part of a dynamical detection system for high-throughput drug screening. It can be expected that with the development of this technology, kidney-on-a-chip systems can become a powerful tool for the study of renal diseases, and play an important role in drug development.
Brain-on-a-chip
The brain is the most complex organ in animals, serving as the center of the nervous system in a vertebrate’s body. The function of the brain is to exert centralized control over the other organs in a body (54). Disruption and dysfunction of neuronal networks are triggered not only by cell death, but also by alterations in the cellular microenvironment or interactions between various cell types, including neurons, glial cells, neural stem cells, and brain vascular endothelial cells. The mechanisms of degenerative diseases, such as Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS), have been extensively studied in the past few decades, but our understanding of these disorders remains incomplete (55). Recently, brain-on-a-chip and central nervous system (CNS) microfluidic devices, due to their small sizes, transparent materials and high throughput capabilities, have become well-suited to capture the detailed behavior of nervous tissues, and to perform neurotoxicity assessments for the various cell types involved. They also allow for fine spatiotemporal control of biochemical and mechanical stimuli over elements of the cellular environment (56). On the basis of these advantages of microfluidic systems, they have been employed by many researchers in studies of nerve function and diseases, including those involving the blood-brain barrier (BBB), AD, PD, and neuro-cytotoxicity, among many others. First, the BBB has structural properties and specialized functional. To protect the brain from harmful substances entering from the bloodstream, the BBB acts as a selective barrier with low permeability. This property is essential to maintain the normal function of the CNS. The BBB is composed of microvascular endothelium surrounded by pericytes and astrocyte foot processes embedded within a basement membrane (57). The primary goal of developing an in vitro human BBB model is to reproduce the various aspects of the BBB in vivo, similar to those found in the brain microvasculature for BBB permeability evaluation and early stage screening of potential CNS drugs (58-60).
For microfluidic-based BBB models, a semi-permeable membrane coated by extracellular matrix is placed between two microchannels, allowing the media to flow over cultured cells. To online measurement of the transepithelial electrical resistance (TEER) values, the electrode pairs were integrated to both the top and bottom microchannels (61). Ugolini et al. presented a new microfluidic device that included two microstructured layers sandwiching a porous membrane for endothelial blood-brain cell transport studies (Figure 4A) (62). The primary benefits of the BBB microsystem included that it employed standard soft-lithography to fabricate a multi-layer device, it exhibited physiological fluid flow and shear stresses, and electrical resistance monitoring and transport studies demonstrated the formation of a brain endothelial barrier. By using this platform, they demonstrated that Br-bEnd5 cells significantly hindered the transport of molecules (40 and 4 kDa dextran) from the top culture chamber to the bottom collection chamber. Giulia et al. demonstrated a neurovascular unit by co-culturing human cerebral endothelial cells with primary rat astrocytes and neurons together (Figure 4B). The three cell types in the microfluidic chip showed specific type of morphological characteristics and functional properties, and human cerebral microvascular endothelial cells formed monolayers with size-selective permeability, similar to those found in vivo (63).
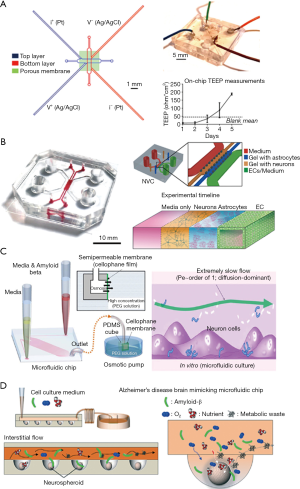
Second, AD is a progressive neurodegenerative disorder, for which the pathological mechanisms are poorly understood. About 70% of the risk is believed to be genetic, with many genes typically involved (64). In recent years, researchers showed that AD is associated with two pathological features—Aβ plaques and neurofibrillary tangles—that are thought to cause brain damage (65). Known as the amyloid-cascade hypothesis, the atypical accumulation of Aβ plaques within the brain is a trigger of AD pathogenesis. The AD research community has adopted this hypothesis as the main explanation for the AD pathological mechanism over the past two decades. However, recently developed Alzheimer’s drugs targeting the removal of Aβ plaques have failed in clinical trials. Thus, there is an urgent need for new in vitro human AD models to help develop a comprehensive understanding of the complex mechanisms of AD pathogenesis, and to screen candidate therapeutic drugs. To overcome some of the limitations of traditional AD systems, several microfluidic systems have been developed in the past decade. Studies of Aβ plaque formation and transmission have been performed by many groups. Choi et al. developed a microfluidic platform capable of generating a gradient of Aβ oligomeric assemblies within microchannels to investigate its neurotoxicity (66). Using an osmotic pump, they induced a slow flow similar to the interstitial flow in the platform (Figure 4C). The neurotoxicity was evaluated by exposing neuronal cells to a gradient of Aβ plaques. Park et al. developed a microfluidic chip containing 3D-neurospheroids that more closely mimicked the in vivo brain microenvironment by providing a constant fluid flow in the interstitial spaces of the brain (Figure 4D) (67). By using this platform, the effect of fluid flow on the neural network, 3D-neurospheroid proliferation, and neural differentiation were investigated. Compared with Aβ treatment under static conditions, treatment with interstitial flow in the microfluidic brain system reduced the viability of neurospheroids and caused significantly more destruction of neural networks.
Third, few realistic PD have been developed so far. Some of the currently available models include the one by Freundt et al., in which they demonstrated a culture model of primary neurons in the presence of fibrils of α-Syn in microfluidic devices with microgrooves (68). They showed that α-Syn fibrils were internalized by neurons and transported along the axons. To make these models more relevant to PD pathophysiology, one could add novel microfluidic features, such as the integration of microvalves to control fluid routing, and the reconstitution of paracrine signaling between the various neural cells, as well as integrating PD patient-derived cells.
There is no doubt that an integrated model of the brain would be a tremendous aid to our understanding of the pathogenesis of brain-related diseases such as AD, PD, and HD. To further investigate neurodegenerative disorders and develop in vitro models that could be applied to drug screening, we believe that it is necessary to combine biological mechanistic and process-oriented knowledge along with the fabrication of appropriate microfluidic devices to reconstitute the microenvironment of the brain. In particular, because the development of brain-related diseases is associated with the dysfunction of other organs, the integration of multiple organs with the brain on a single microdevice is a trend of the future. For drug-screening studies, a high-throughput device must be engineered to be stable and low-cost. In the future, the combination of brain organoids with a high-throughput microfluidic device could open new doors for the basic study of neuronal development, drug-screening, and tissue-engineering applications.
Multi-organ-on-a-chip
Multi-organ-on-a-chip aims to build a highly functional and robust system to mimic the whole-body and simulate human functional responses and interaction with one another, which integrate several artificial organs into a single device via internal or external recirculation fluidic flow. In this system, a variety of cells derived from different organs and tissues are cultured on the chip in a physiological manner in respective culture chambers. For example, Fan and his colleagues reported a laminated microfluidic device for comprehensive preclinical testing in the drug ADME process (absorption, metabolism, distribution and excretion), which integrated intestine, liver, tumor, lung, heart and fat into a single microdevice (9). The pharmacokinetic parameters of propranolol, thiopentone and pentobarbital was successfully conducted. In another example, Shuler et al. reported that 14 chambers (13 organs) were arranged in the chip in proportion to the body, which could be used to simulate drug distribution, metabolism, and action in the body (11). It is based upon a physiologically based pharmacokinetic–pharmacodynamics model, where multiple chambers representing different organs are connected with fluidic channels to mimic multi-organ interactions within the body. Clearly, multi-organ-on-a-chips provide more pharmacokinetic properties and toxic effects between each organ, thus being more suitable for drug testing. It is foreseeable that multi-organ chips are a significant area of future drug toxicity research.
Conclusions
In this review, organ-on-a-chip devices that contain critical microarchitecture, spatiotemporal cell-cell interactions, and extracellular microenvironments have been discussed. We reviewed recent works on heart-, liver-, kidney-, and brain-on-a-chip systems for drug toxicity testing, as well as multi-organ-on-a-chip for drug toxicity screening. Significant progress has been achieved to date, even though some factors limiting the development of organ-on-chips devices remain, including those related with cell sources, materials, and technologies. Organ-on-a-chip technologies can overcome many of the shortcomings of traditional two-dimensional cell culture models and animal experiments. In addition, this approach has the potential to establish highly biomimetic in vitro physiology models to bridge the gap between preclinical and clinical outcomes, which may even affect the development of the pharmaceutical industry. Further, the ultimate goal of a “human-on-chip” that integrates all of the important organs, and could potentially replace animal experiments in comprehensive drug toxicity studies, is now conceivable.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 21675017), the National Key Research and Development Program of China (No. SQ2017YFC170204-001), and the Fundamental Research Funds for the Central Universities, China (No. DUT17LK25).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/mps.2018.09.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zheng F, Fu F, Cheng Y, et al. Organ-on-a-Chip Systems: Microengineering to Biomimic Living Systems. Small 2016;12:2253-82. [Crossref] [PubMed]
- Balijepalli A, Sivaramakrishan V. Organs-on-chips: research and commercial perspectives. Drug Discov Today 2017;22:397-403. [Crossref] [PubMed]
- Kim S, Kim HJ, Jeon NL. Biological applications of microfluidic gradient devices. Integr Biol (Camb) 2010;2:584-603. [Crossref] [PubMed]
- Akbari E, Spychalski GB, Rangharajan KK, et al. Flow dynamics control endothelial permeability in a microfluidic vessel bifurcation model. Lab Chip 2018;18:1084-93. [Crossref] [PubMed]
- Ma X, Qu X, Zhu W, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A 2016;113:2206-11. [Crossref] [PubMed]
- Maschmeyer I, Lorenz AK, Schimek K, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015;15:2688-99. [Crossref] [PubMed]
- Huh D, Torisawa Y, Hamilton GA, et al. Microengineered physiological biomimicry: Organs-on-Chips. Lab Chip 2012;12:2156-64. [Crossref] [PubMed]
- Materne E, Tonevitsky AG, Marx U. Chip-based liver equivalents for toxicity testing - organotypicalness versus cost-efficient high throughput. Lab Chip 2013;13:3481-95. [Crossref] [PubMed]
- An F, Qu Y, Luo Y, et al. A Laminated Microfluidic Device for Comprehensive Preclinical Testing in the Drug ADME Process. Sci Rep 2016;6:25022. [Crossref] [PubMed]
- Ataç B, Wagner I, Horland R, et al. Skin and hair on-a-chip: in vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip 2013;13:3555-61. [Crossref] [PubMed]
- Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol Bioeng 2016;113:2213-27. [Crossref] [PubMed]
- Grosberg A, Alford PW, McCain ML, et al. Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip 2011;11:4165. [Crossref] [PubMed]
- Agarwal A, Goss JA, Cho A, et al. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013;13:3599-608. [Crossref] [PubMed]
- McCain ML, Agarwal A, Nesmith HW, et al. Micromolded gelatin hydrogels for extended culture of engineered cardiac tissues. Biomaterials 2014;35:5462-71. [Crossref] [PubMed]
- Nawroth JC, Scudder LL, Halvorson RT, et al. Automated fabrication of photopatterned gelatin hydrogels for organ-on-chips applications. Biofabrication 2018;10:025004 [Crossref] [PubMed]
- Stancescu M, Molnar P, McAleer CW, et al. A phenotypic in vitro model for the main determinants of human whole heart function. Biomaterials 2015;60:20-30. [Crossref] [PubMed]
- Lind JU, Yadid M, Perkins I, et al. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip 2017;17:3692-703. [Crossref] [PubMed]
- Tanaka Y, Sato K, Shimizu T, et al. A micro-spherical heart pump powered by cultured cardiomyocytes. Lab Chip 2007;7:207-12. [Crossref] [PubMed]
- Sidorov VY, Samson PC, Sidorova TN, et al. I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater 2017;48:68-78. [Crossref] [PubMed]
- Schroer AK, Shotwell MS, Sidorov VY, et al. I-Wire Heart-on-a-Chip II: Biomechanical analysis of contractile, three-dimensional cardiomyocyte tissue constructs. Acta Biomater 2017;48:79-87. [Crossref] [PubMed]
- Wang G, McCain ML, Yang L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014;20:616-23. [Crossref] [PubMed]
- Marsano A, Conficconi C, Lemme M, et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016;16:599-610. [Crossref] [PubMed]
- Parsa H, Wang BZ, Vunjak-Novakovic G. A microfluidic platform for the high-throughput study of pathological cardiac hypertrophy. Lab Chip 2017;17:3264-71. [Crossref] [PubMed]
- Kobuszewska A, Tomecka E, Zukowski K, et al. Heart-on-a-Chip: An Investigation of the Influence of Static and Perfusion Conditions on Cardiac (H9C2) Cell Proliferation, Morphology, and Alignment. SLAS Technol 2017;22:536-46. [Crossref] [PubMed]
- Zhang YS, Arneri A, Bersini S, et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016;110:45-59. [Crossref] [PubMed]
- Wang Z, Lee SJ, Cheng H, et al. 3D bioprinted functional and contractile cardiac tissue constructs. Acta Biomater 2018;70:48-56. [Crossref] [PubMed]
- Zuchowska A, Kwapiszewska K, Chudy M, et al. Studies of anticancer drug cytotoxicity based on long-term HepG2 spheroid culture in a microfluidic system. Electrophoresis 2017;38:1206-16. [Crossref] [PubMed]
- Bi Y, Kazolias D, Duignan DB. Use of cryopreserved human hepatocytes in sandwich culture to measure hepatobiliary transport. Drug Metab Dispos 2006;34:1658-65. [Crossref] [PubMed]
- Godoy P, Hewitt NJ, Albrecht U, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013;87:1315-530. [Crossref] [PubMed]
- Du C, Narayanan K, Leong MF, et al. Induced pluripotent stem cell-derived hepatocytes and endothelial cells in multi-component hydrogel fibers for liver tissue engineering. Biomaterials 2014;35:6006-14. [Crossref] [PubMed]
- Sison-Young RL, Lauschke VM, Johann E, et al. A multicenter assessment of single-cell models aligned to standard measures of cell health for prediction of acute hepatotoxicity. Arch Toxicol 2017;91:1385-400. [Crossref] [PubMed]
- Hendriks DFG, Fredriksson Puigvert L, Messner S, et al. Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Sci Rep 2016;6:35434. [Crossref] [PubMed]
- Leite SB, Roosens T, El Taghdouini A, et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 2016;78:1-10. [Crossref] [PubMed]
- Rennert K, Steinborn S, Gröger M, et al. A microfluidically perfused three dimensional human liver model. Biomaterials 2015;71:119-31. [Crossref] [PubMed]
- Bell CC, Hendriks DF, Moro SM, et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 2016;6:25187. [Crossref] [PubMed]
- Weng YS, Chang SF, Shih MC, et al. Scaffold-Free Liver-On-A-Chip with Multiscale Organotypic Cultures. Adv Mater 2017;29: [Crossref] [PubMed]
- Bavli D, Prill S, Ezra E, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A 2016;113:E2231-40. [Crossref] [PubMed]
- Bhise NS, Manoharan V, Massa S, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016;8:014101 [Crossref] [PubMed]
- Redfern WS, Bialecki R, Ewart L, et al. Impact and prevalence of safety pharmacology-related toxicities throughout the pharmaceutical life cycle. J Pharmacol Tox Met 2010;62:e29 [Crossref]
- Chu X, Bleasby K, Evers R. Species differences in drug transporters and implications for translating preclinical findings to humans. Expert Opin Drug Met 2012;9:237-52. [Crossref] [PubMed]
- Orosz DE, Woost PG, Kolb RJ, et al. Growth, immortalization, and differentiation potential of normal adult human proximal tubule cells. In Vitro Cell Dev Biol Anim 2004;40:22-34. [Crossref] [PubMed]
- Jang KJ, Mehr AP, Hamilton GA, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol (Camb) 2013;5:1119-29. [Crossref] [PubMed]
- Weber EJ, Chapron A, Chapron BD, et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int 2016;90:627-37. [Crossref] [PubMed]
- Musah S, Mammoto A, Ferrante TC, et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat Biomed Eng 2017;1: [Crossref] [PubMed]
- Mu X, Zheng W, Xiao L, et al. Engineering a 3D vascular network in hydrogel for mimicking a nephron. Lab Chip 2013;13:1612-8. [Crossref] [PubMed]
- Nieskens TT, Wilmer MJ. Kidney-on-a-chip technology for renal proximal tubule tissue reconstruction. Eur J Pharmacol 2016;790:46-56. [Crossref] [PubMed]
- Jang K, Cho HS, Kang DH, et al. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr Biol 2011;3:134-41. [Crossref] [PubMed]
- Sakolish CM, Mahler GJ. A novel microfluidic device to model the human proximal tubule and glomerulus. RSC Adv 2017;7:4216-25. [Crossref]
- Ferrell N, Ricci KB, Groszek J, et al. Albumin handling by renal tubular epithelial cells in a microfluidic bioreactor. Biotechnol Bioeng 2012;109:797-803. [Crossref] [PubMed]
- Snouber LC, Letourneur F, Chafey P, et al. Analysis of transcriptomic and proteomic profiles demonstrates improved Madin-Darby canine kidney cell function in a renal microfluidic biochip. Biotechnol Progr 2012;28:474-84. [Crossref] [PubMed]
- Baudoin R, Griscom L, Monge M, et al. Development of a renal microchip for in vitro distal tubule models. Biotechnol Prog 2007;23:1245-53. [PubMed]
- Nieskens TT, Peters JG, Schreurs MJ, et al. A Human Renal Proximal Tubule Cell Line with Stable Organic Anion Transporter 1 and 3 Expression Predictive for Antiviral-Induced Toxicity. AAPS J 2016;18:465-75. [Crossref] [PubMed]
- Singh M, Tong Y, Webster K, et al. 3D printed conformal microfluidics for isolation and profiling of biomarkers from whole organs. Lab Chip 2017;17:2561-71. [Crossref] [PubMed]
- Pelvig DP, Pakkenberg H, Stark AK, et al. Neocortical glial cell numbers in human brains. Neurobiol Aging 2008;29:1754-62. [Crossref] [PubMed]
- Osaki T, Shin Y, Sivathanu V, et al. In Vitro Microfluidic Models for Neurodegenerative Disorders. Adv Healthc Mater 2018;7:1700489 [Crossref] [PubMed]
- van den Berg A, Craighead HG, Yang P. From microfluidic applications to nanofluidic phenomena. Chem Soc Rev 2010;39:899-900. [Crossref] [PubMed]
- Brown JA, Codreanu SG, Shi M, et al. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J Neuroinflammation 2016;13:306. [Crossref] [PubMed]
- Ogunshola OO. In vitro modeling of the blood-brain barrier: simplicity versus complexity. Curr Pharm Des 2011;17:2755-61. [Crossref] [PubMed]
- Nakagawa S, Deli MA, Kawaguchi H, et al. A new blood–brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem Int 2009;54:253-63. [Crossref] [PubMed]
- Nicolazzo JA, Charman SA, Charman WN. Methods to assess drug permeability across the blood-brain barrier. J Pharm Pharmacol 2006;58:281-93. [Crossref] [PubMed]
- Griep LM, Wolbers F, de Wagenaar B, et al. BBB ON CHIP: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices 2013;15:145-50. [Crossref] [PubMed]
- Ugolini GS, Occhetta P, Saccani A, et al. Design and validation of a microfluidic device for blood-brain barrier monitoring and transport studies. J Micromech Microeng 2018;28: [Crossref]
- Adriani G, Ma D, Pavesi A, et al. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab Chip 2017;17:448-59. [Crossref] [PubMed]
- Ballard C, Gauthier S, Corbett A, et al. Alzheimer's disease. Lancet 2011;377:1019-31. [Crossref] [PubMed]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002;297:353-6. [Crossref] [PubMed]
- Choi YJ, Chae S, Kim JH, et al. Neurotoxic amyloid beta oligomeric assemblies recreated in microfluidic platform with interstitial level of slow flow. Sci Rep 2013;3:1921. [Crossref] [PubMed]
- Park J, Lee BK, Jeong GS, et al. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer's disease. Lab Chip 2015;15:141-50. [Crossref] [PubMed]
- Freundt EC, Maynard N, Clancy EK, et al. Neuron-to-neuron transmission of alpha-synuclein fibrils through axonal transport. Ann Neurol 2012;72:517-24. [Crossref] [PubMed]
Cite this article as: Deng J, Qu Y, Liu T, Jing B, Zhang X, Chen Z, Luo Y, Zhao W, Lu Y, Lin B. Recent organ-on-a-chip advances toward drug toxicity testing. Mesentery Peritoneum 2018;2:8.


